Similarity and uncertainty in medical image analysis
Image similarity is one of the most integral parts of medical image computing. Establishing correspondences across medical scans of different patients, time-points or modalities is key to numerous medical image analysis applications such as: atlas-based segmentation, motion estimation, longitudinal studies and multi-modal fusion. Defining similarity across images is challenging due to global and local changes in image contrast, noise and the variety of physical principals, which are used to acquire different modalities. One aim of this project is to improve or develop novel similarity metrics, which are invariant to modality and robust against contrast variations and noise while still being very discriminative for important anatomical or geometric features.
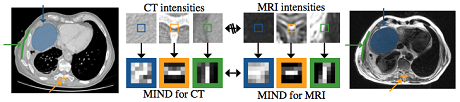
The modality independent neighbourhood descriptor (MIND) [1] is a multi-dimensional local image descriptor (see Fig. 1), which we have developed for multi-modal registration based on the concept of self-similarities. We have demonstrated its improvements over state-of-the-art methods such as mutual information for the alignment of thoracic CT and MRI scans of patients with empyema, a lung disease. It can also be used for registering scans of the same modality, e.g. 4DCT scans, where it has been shown to improve accuracy and robustness. Each MIND descriptor is calculated based on patch distances (within the local neighbourhood of the same scan). Comparison of MIND representations is performed as simple sum of squared/absolute differences of its entries. The self-similarity context (SSC) [2] is an improvement of MIND, which redefines the neighbourhood layout to improve the robustness of the matching and better describe the contextual information. We have derived an efficient quantisation scheme, which enables a very fast evaluation of pair-wise distances using the Hamming weight and shown that is applicable to register challenging image modalities such as MRI and ultrasound.
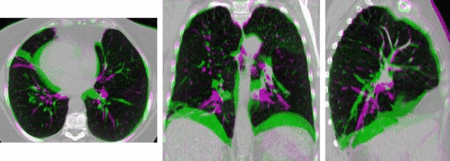
Uncertainty is inherently an important issue to all image analysis tasks. However, few common algorithms estimate a measure of uncertainty for the calculated results. Automatic error detection would be very valuable in the practical use of medical image analysis tools, as it could provide the clinician with more confidence on whether or not to use a computer-generated analysis. Discrete optimisation techniques (based on Markov random field models) may offer useful solutions to determine uncertainty, because they can be used to infer probabilistic estimates over a flexible range of model parameters. For the application of image registration, we have developed a framework that optimises over a very large space of potential displacements (dense displacement sampling, deeds) [3,4]. It enables very fast and accurate motion estimation and has been evaluated on a large set of lung CT scans. An example outcome of a registration is shown in Fig. 2 (quantitative results can be found at http://empire10.isi.uu.nl/res_deedsmind.php). The uncertainty over all potential motion parameters can be calculated using well-known message passing algorithms and enable us to quantify local registration accuracy. This has been used in [5] to improve segmentation accuracy of MRI brain scans. Not only the displacement vectors contain uncertainties, but also the underlying image representation. In practice, a transformation model parameterised by uniform B-spline control-points is often used. We have presented a more flexible image model in [6], consisting of multiple complementary layers of supervoxels. Estimating the best local image model and determining its trustworthiness forms another part of the current work in this project.
Software developed by M.P. Heinrich for the publications below, can be freely downloaded from: www.mpheinrich.de/software.html
This includes a very efficient and accurate 3D single- and multimodal registration package deeds and reference code to obtain MIND and SSC descriptors.
Selected Publications
- Mattias P. Heinrich, Mark Jenkinson, Manav Bhushan, Tahreema Matin, Fergus V. Gleeson, Sir Michael Brady, Julia A. Schnabel.
MIND: Modality Independent Neighbourhood Descriptor for Multi-modal Deformable Registration.
Medical Image Analysis. vol. 16(7) Oct. 2012, pp. 1423–1435 - Mattias Paul Heinrich, Mark Jenkinson, Bartlomiej W. Papiez, Sir Michael Brady, Julia A. Schnabel.
Towards Realtime Multimodal Fusion for Image-Guided Interventions Using Self-similarities
In: Medical Image Computing and Computer-Assisted Intervention – MICCAI 2013, Lecture Notes in Computer Science Volume 8149, 2013, pp 187-194 - Mattias P. Heinrich, Mark Jenkinson, Sir Michael Brady, Julia A. Schnabel.
MRF-based Deformable Registration and Ventilation Estimation of Lung CT.
IEEE Transaction on Medical Imaging. Vol. 32(7), 1239 - 1248, 2013 - MP Heinrich, M Jenkinson, M Brady, JA Schnabel.
Globally Optimal Registration on a Minimum Spanning Tree using Dense Displacement Sampling
In: Medical Image Computing and Computer Assisted Intervention (MICCAI) 2012. LNCS 7512, pp. 115-122. Springer, Berlin (2012) - Mattias P. Heinrich, Ivor J.A. Simpson, Mark Jenkinson, Sir Michael Brady, Julia A. Schnabel.
Uncertainty Estimates for Improved Accuracy of Registration-Based Segmentation Propagation using Discrete Optimisation
MICCAI Workshop on Segmentation, Algorithms, Theory and Applications (SATA), Nagoya 2013 - Mattias P. Heinrich, Mark Jenkinson, Bartlomiej W. Papiez, Fergus V. Gleeson, Sir Michael Brady, Julia A. Schnabel.
Edge- and Detail-Preserving Sparse Image Representations for Deformable Registration of Chest MRI and CT Volumes.
In: Information Processing in Medical Imaging (IPMI) 2013. LNCS 7917, 463-474, Springer (2013)
Project Team
Jun.-Prof. Dr. Mattias P. Heinrich
Dr. Jan Ehrhardt
Prof. Dr. Heinz Handels
Cooperation Partners
Prof. Dr. Julia A. Schnabel
Institute of Biomedical Engineering
University of Oxford

- Research
- AI und Deep Learning in Medicine
- Medical Image Processing and VR-Simulation
- Integration and Utilisation of Medical Data
- Sensor Data Analysis for Assistive Health Technologies
- Medical Image Computing and Artificial Intelligence
- Medical Data Science Lab
- Medical Deep Learning Lab
- Junior Research Group Diagnostics and Research of Movement Disorders
- Former Medical Data Engineering Lab
